Running Python code requires a running Python kernel. Click the –> Live Code button above on this page to run the code below.
Warning
🚧 This site is under construction! As of now, the Python kernel may not run on the page or have very long wait times. Also, expect typos.👷🏽♀️
Example: Supervised Classification App#
A supervised classification method fits the project requirements well and is so a good place to start. The nature of your Data and organizational needs dictate which methods you can use. So what type of data works with supervised classification methods?
One of the features (columns) contains mutually exclusive categories you want to predict (the dependent variable).
At least one other feature (the independent variable(s)).
This will be a simple example. Simple data. Simple model. Simple interface. However, it does demonstrate the minimum requirements for part C. We’ll also show how things can progressively be improved, building on the working code. Simple is a great place to start -scaling up is typically easier than going in the other direction.
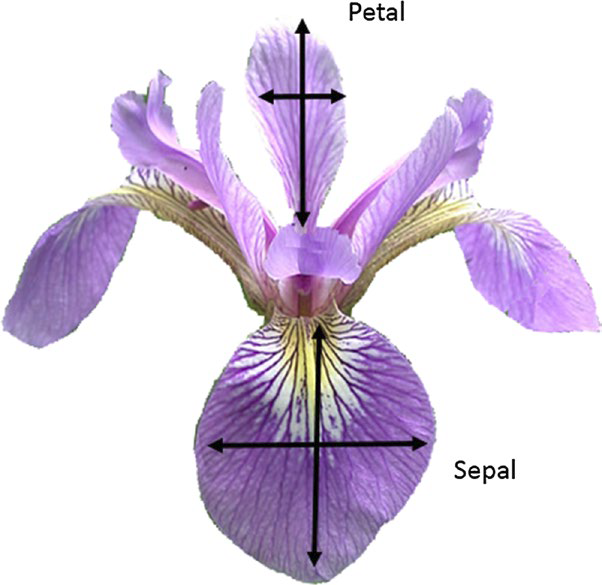
Let’s look at the famous Fisher’s Iris data set:
| sepal-length | sepal-width | petal-length | petal-width | type | |
|---|---|---|---|---|---|
| 0 | 5.1 | 3.5 | 1.4 | 0.2 | Iris-setosa |
| 1 | 4.9 | 3.0 | 1.4 | 0.2 | Iris-setosa |
| 2 | 4.7 | 3.2 | 1.3 | 0.2 | Iris-setosa |
| 3 | 4.6 | 3.1 | 1.5 | 0.2 | Iris-setosa |
| 4 | 5.0 | 3.6 | 1.4 | 0.2 | Iris-setosa |
| ... | ... | ... | ... | ... | ... |
| 145 | 6.7 | 3.0 | 5.2 | 2.3 | Iris-virginica |
| 146 | 6.3 | 2.5 | 5.0 | 1.9 | Iris-virginica |
| 147 | 6.5 | 3.0 | 5.2 | 2.0 | Iris-virginica |
| 148 | 6.2 | 3.4 | 5.4 | 2.3 | Iris-virginica |
| 149 | 5.9 | 3.0 | 5.1 | 1.8 | Iris-virginica |
Though we described everything as “simple,” we’ll also see that this dataset is quite rich with angles to investigate. At this point, we have many options, but for a classification project we need a categorical feature as our dependent variable, and for this, we only have the choice: type.
| sepal-length | sepal-width | petal-length | petal-width | type | |
|---|---|---|---|---|---|
| 0 | 5.1 | 3.5 | 1.4 | 0.2 | Iris-setosa |
| 1 | 4.9 | 3.0 | 1.4 | 0.2 | Iris-setosa |
| 2 | 4.7 | 3.2 | 1.3 | 0.2 | Iris-setosa |
| 3 | 4.6 | 3.1 | 1.5 | 0.2 | Iris-setosa |
| 4 | 5.0 | 3.6 | 1.4 | 0.2 | Iris-setosa |
| ... | ... | ... | ... | ... | ... |
| 145 | 6.7 | 3.0 | 5.2 | 2.3 | Iris-virginica |
| 146 | 6.3 | 2.5 | 5.0 | 1.9 | Iris-virginica |
| 147 | 6.5 | 3.0 | 5.2 | 2.0 | Iris-virginica |
| 148 | 6.2 | 3.4 | 5.4 | 2.3 | Iris-virginica |
| 149 | 5.9 | 3.0 | 5.1 | 1.8 | Iris-virginica |
The highlighted column, type provides a category to predict/classify (dependent variables), and the non-highlighted columns are something by which to make that prediction/classification (independent variables).